

The results from PRANK show that the alignments of the internal repeats are similar to those done manually, suggesting multiple feasible alignments for some regions. The bioinformatic program, PRANK, was used because it was designed to align sequences with large gaps and indels. Furthermore, because two different manual alignments of the SpTrf sequences are feasible because of the multiple internal repeats, it is not known whether additional alternative alignments can be identified using different approaches. Because manual alignments are time consuming for evaluating newly available SpTrf sequences, computational approaches were evaluated for the sequences reported previously. This mosaic pattern necessitates the insertion of large gaps, which has made alignments of the deduced protein sequences computationally difficult such that only manual alignments have been reported previously. The gene sequences have a series of internal repeats in a mosaic pattern that is characteristic of this family.
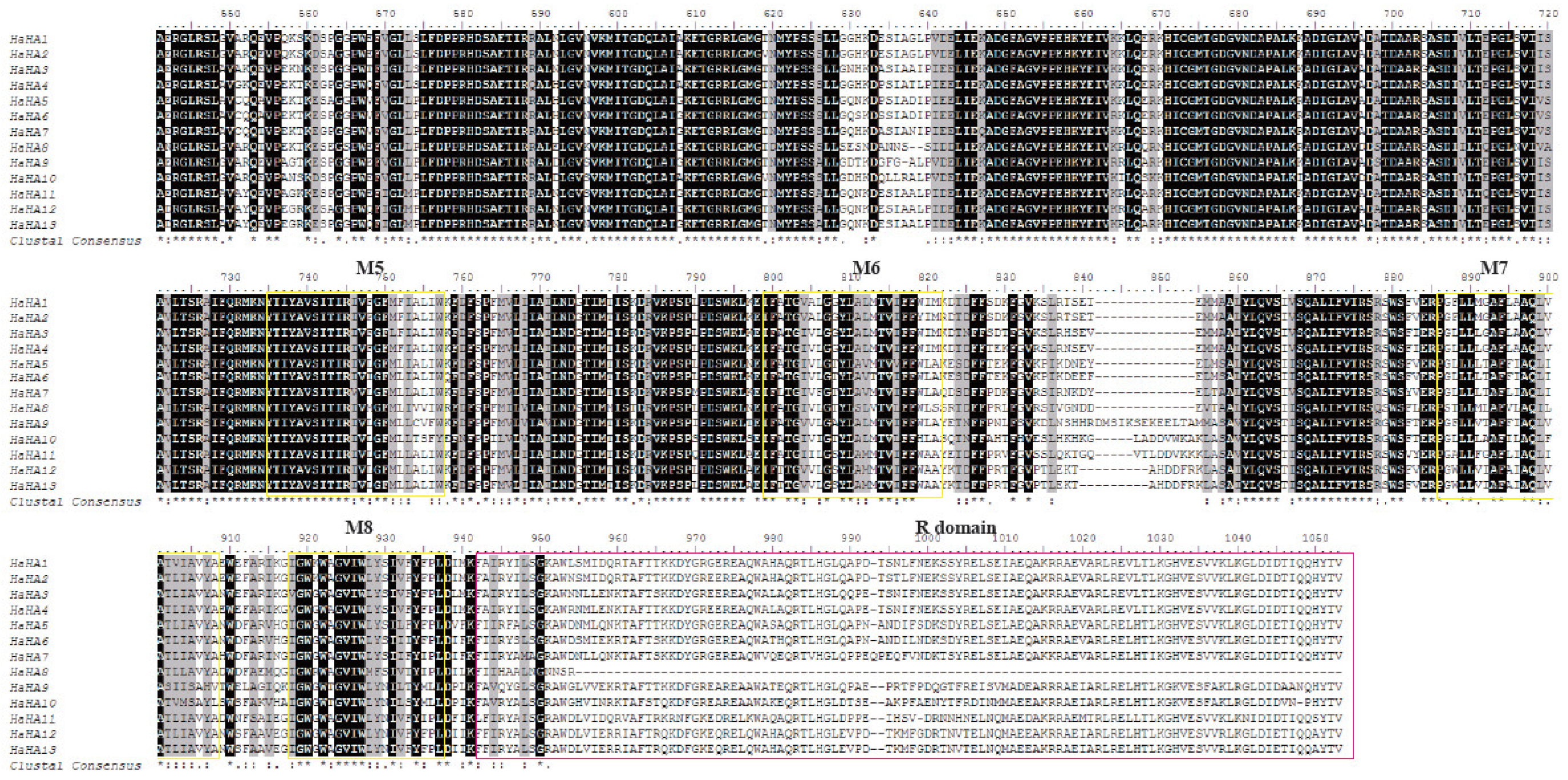
The SpTransformer ( SpTrf) gene family encodes a set of proteins that function in the sea urchin immune system. Department of Biological Sciences, George Washington University, Washington, DC, United States.


 0 kommentar(er)
0 kommentar(er)
